About me
G’day! My name is Jaimi Gray and I am an evolutionary biologist who specialises in 3D imaging. I am currently working remotely from Australia on the oVert project for the Florida Museum of Natural History at the University of Florida.
How did I get here?
It all began in 2013 when I started a research project at the South Australian Museum. I opened my very first CT scan—the skull of an Australian mountain dragon (Rankinia diemensis). It was love at first sight, and from that fateful moment onward my life would revolve around working with CT data. Since then I have been CT scanning and reconstructing animal specimens in 3D, and using the data to build 3D models for morphological descriptions, shape analysis, and teaching resources.
I was born and bred in rural South Australia. I spent the first 18 years of my life on a sheep and cattle farm near a small country town called Millicent. In 2010, I left my hometown to pursue a degree in Evolutionary Biology, majoring in palaeontology, at the University of Adelaide. My first experience with CT data was for a research project in the final year of my degree, where I used scans and 3D models to study the morphology of two closely related dragon lizard species.
Upon completion of my undergraduate degree, I undertook a year of honours, during which I CT scanned fossil jaw bones and extant dragon lizard specimens in an attempt to identify fossil specimens in South Australian cave systems. During this project, it became painfully apparent to me that more answers were needed about the extant animals before substantial claims could be made about these dragon fossils.
Wanting to solve more dragon mysteries, I went on to complete my PhD at the University of Adelaide on Australian dragon lizard skull morphology in 2018. I loved the dragons and had grown to love all reptiles and the herpetological community in Australia; but by the time I finished my PhD I had realised my true passion lay in the methods I had been using during the last five years. I loved 3D imaging and CT scanning, and everything that came along with it.
 3D model made from the CT scan of the skull of one of Australia’s iconic dragon lizards, Moloch horridus (the thorny devil), overlayed on an image of the animal in life.
3D model made from the CT scan of the skull of one of Australia’s iconic dragon lizards, Moloch horridus (the thorny devil), overlayed on an image of the animal in life.I started working on other things. I jumped from vertebrates to invertebrates when I landed my first postdoctoral position at the University of Auckland in New Zealand. Here I used CT data to study the weapon morphology of New Zealand’s endemic harvestmen (a type of arachnid). Up until this point I had worked with hard tissues (skeletons of vertebrates and exoskeletons of invertebrates), and I had already decided that I wanted to level up my 3D imaging game by adding soft tissue imaging techniques into my skillset.
 Diagram of the CT scan of a New Zealand Neopilionid harvestman, showing examples of an X-ray projection, reconstructed CT slice (cross-section), and 3D model made from CT scan.
Diagram of the CT scan of a New Zealand Neopilionid harvestman, showing examples of an X-ray projection, reconstructed CT slice (cross-section), and 3D model made from CT scan.For my second postdoctoral position, I applied and was awarded an Australian Government Funded scholarship called the Endeavour Research Leadership Award. I moved to Tulsa, Oklahoma, where I learned contrast-enhanced staining techniques for CT scanning soft tissue anatomy of animals and started my research on the neuroanatomy of terrestrial and marine elapid snakes. At the end of this experience, I was proud of how far I had come. Now, I can use CT imaging to reconstruct 3D models of anything. Fossil or living. Vertebrate or invertebrate. Hard tissue or soft tissue. And I wanted to 3D image all of the things.
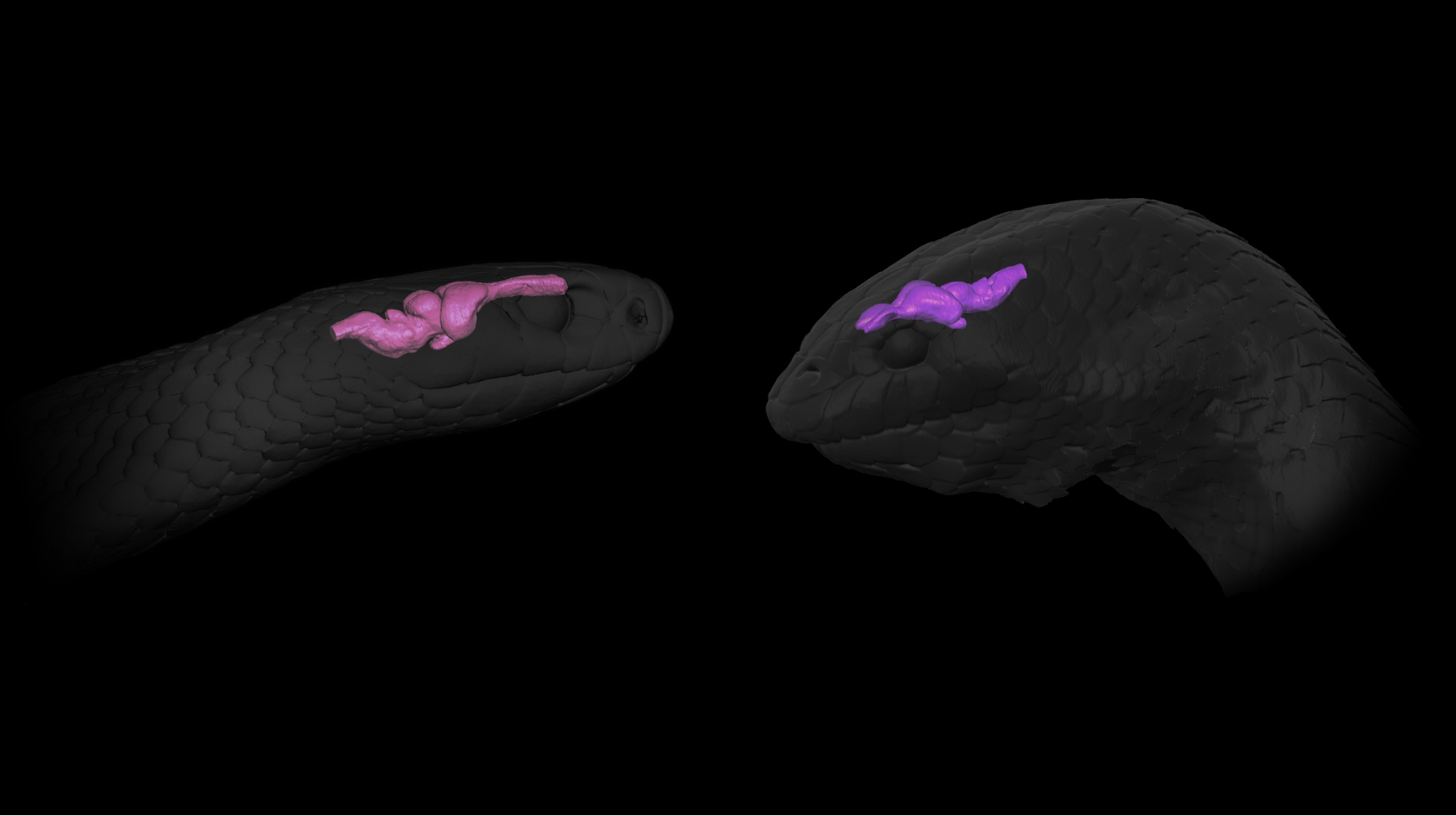 3D models of the head and brains of a land snake (Pseudonaja textilis), and a sea snake (Aipysurus laevis), produced from DiceCT scans.
3D models of the head and brains of a land snake (Pseudonaja textilis), and a sea snake (Aipysurus laevis), produced from DiceCT scans.Scan all the Vertebrates!
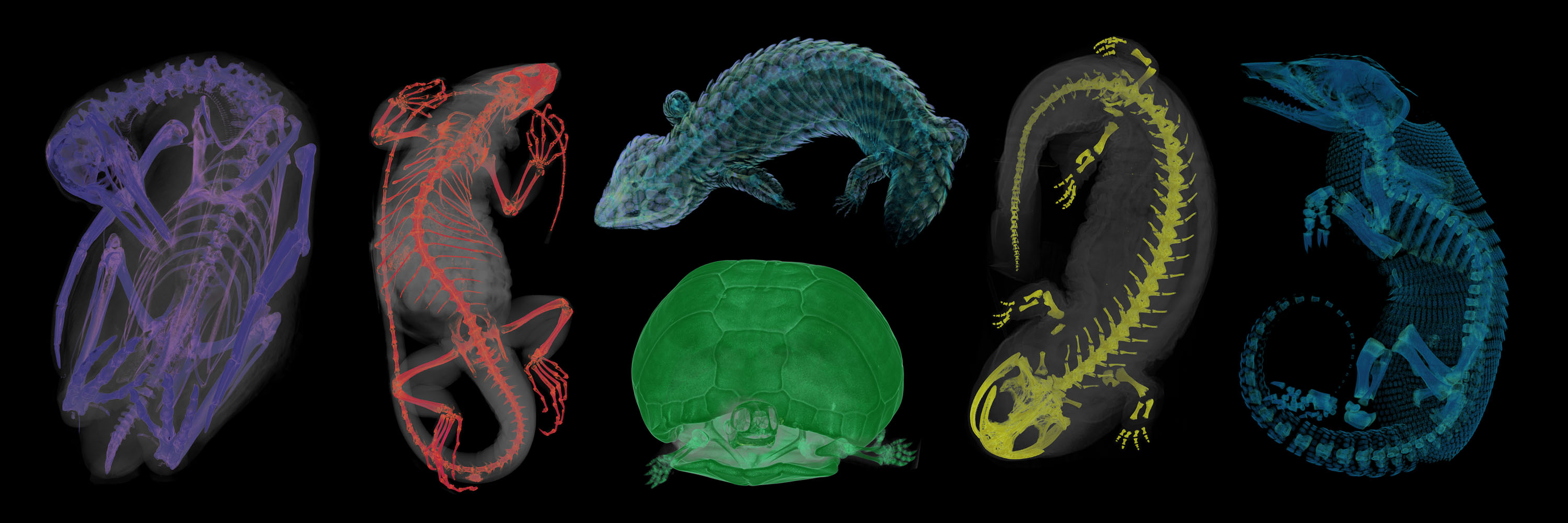
At the beginning of 2019, along came the perfect opportunity. I joined the team at Florida Museum of Natural History, who are working on the NSF-funded oVert (“open vertebrate”) project. The oVert team, along with more than 20 other collaborating institutions, aims to facilitate exploration of vertebrate diversity by doing just that—CT scanning huge numbers of natural history specimens. Even better, we are making these scans publicly available using the MorphoSource digital repository. So far, we have scanned a massive 8,300 specimens representing approximately 4,300 genera in over 700 families of vertebrates, from around 50 different natural history collections. As part of the oVert project I have been using CT data to produce Sketchfab models as teaching resources for vertebrate evolution and comparative anatomy. A major project of mine has been creating a collection of models that demonstrate the anatomy and homology of the vertebrate forelimb. My Sketchfab models have already been used in several evolution and comparative anatomy university courses. Now I am working on more teaching collections, including a collection of dentition models.
My workflow
For those unfamiliar, CT scanning begins with a series of X-ray images of a specimen, all taken from different angles. We can use these X-ray images to reconstruct the dense tissue of an animal in 3D—this is the Computed Tomography or CT part of the process. The resulting data set is an “image stack”. Each individual image or “slice” in the stack represents a cross-section through the specimen, and together the images make up a 3D volume, complete with internal structures.
 3D models of the head, brain, and trigeminal nerves of the olive sea snake (Aipysurus laevis) with cross section of the head, produced from a DiceCT scan of a specimen from the Field Museum of Natural History.
3D models of the head, brain, and trigeminal nerves of the olive sea snake (Aipysurus laevis) with cross section of the head, produced from a DiceCT scan of a specimen from the Field Museum of Natural History.Just as any 2D image is made up of pixels, so too are 3D volumes made up of voxels. To make 3D meshes that can be made into Sketchfab models, I need to go through the CT slices and use different tools to identify the voxels that will make up the model. This part of the process is essentially a digital dissection, where I extract the structures I want without ever having to touch a scalpel. Once I have identified the material I want to make up the model, I can generate mesh files.
Digital dissections can be very time-consuming. Typically, skeletal tissue and other hard material is easily visualised using density thresholds, but it gets difficult when we want to visualise individual bones and need to separate them from one another. While the soft tissue anatomy is visible in a typical CT scan, you cannot distinguish between different types of soft tissue. I use a staining technique called diffusible iodine-based contrast-enhanced CT, also known as DiceCT. The staining protocol 1) increases the radiodensity, and therefore the visibility of soft tissue in CT scans, and 2) enhances the contrast between different types of tissue. With DiceCT, I can use CT data sets to build 3D models of the soft tissue anatomy of a specimen.
How I use Sketchfab
Since my models are scientific ones, I like the scenes I create to simply and visually convey a wealth of information about the system they represent. To achieve this, I use background images that contain colour keys and other information about the model the viewer is seeing. My models are made up of multiple mesh files, and this allows me to individually colour elements to match the colour key, and to make different parts of the model transparent so that other parts inside can be seen. These visualisations, along with annotations added to the model itself, allow me to visually communicate information about the model with the viewer.
I’ve also been combining multiple models to create large Sketchfab scenes. This allows the viewer to see multiple models at a time and make comparisons among them. I add information about how the models relate to one another—I achieve this by using mesh editing software to create additions such as 3D arrows or 3D evolutionary trees. Lastly, I use the annotations to add links to each model in the scene, which take the viewer to a full resolution, individual version of that model that can be explored in more detail. The resulting scenes are information rich and allow users to take a virtual tour through a (sometimes complex) evolutionary system.
Which Sketchfab features I find most useful
The thing I love most about Sketchfab is that it enhances my 3D models. It has allowed me to create models that are more than just an OBJ or an STL. The ability to create an entire 3D scene using backgrounds, different rendering styles within one scene, annotations, and virtual tours around the models, results in an intuitive and information-rich method of communicating in three dimensions. I have been making 3D models for a long time, but am relatively new to Sketchfab. My work on the oVert project brought me headfirst into the Sketchfab world, and now I use it extensively for both the oVert project and other research I am involved with.
Sketchfab really enhances the accessibility of 3D models. For non-experts, finding your way around CT data can be difficult. But with Sketchfab, I can share ready-made 3D models with other scientists, students, and the public, without them needing any special software or 3D modelling expertise—just the link and an internet browser! I have mostly been publishing 3D models through the Blackburn Lab Sketchfab page. I do have my own Sketchfab page with several models and many under construction, but these can’t be seen by the public yet as they are associated with scientific research that is yet to be published—watch this space!
If you want to follow updates on my work, you can follow me on Twitter, keep an eye on the Blackburn Lab Sketchfab Page, or my own Sketchfab page.